This web page was produced as an assignment for an undergraduate course at Davidson College.
A Review of the paper
“Creating Bacterial Strains from Genomes That Have Been Cloned and Engineered in Yeast” by Lartigue et al.
Overview
The purpose of this paper was to introduce a new experimental method that allows researchers to transplant the genome of one bacterial strain into a eukaryotic model organism like yeast and then transplant it back into a new bacterial strain. What makes this technique so unique is the fact that it allows researchers to make modifications in the bacterial genome, previously impossible, using the vast array of genetic techniques already available for yeast. More specifically, the authors transplant claimed to have transplanted modified and unmodified genome of the organism Mycoplasma mycoides into a new into a relative bacterial organism, namely Mycoplasm capricolum, after cloning it in yeast.
Methodology
The first step in their method involves transforming the M. mycoides genomic DNA with a vector that includes a number of markers that would allow it to be incorporated and to proliferate in the yeast cells. They named the clone of the genomic M. mycoides coupled with the vector YCpMmyc1.1. They then obtained two spheroplast yeast cell lines (VL6-48N and W303a) and transfected it them with YCpMmyc1.1, which they isolated from the M. mycoides. They then took a group of transformed yeast cells and performed a seamless deletion, which is a deletion that leaves a limited amount of foreign DNA in the modified genome, of one of the M. mycoides nonessential genes found in the YCpMmyc1.1. As they mentioned in the paper the technique that they used to make this seamless deletion cannot be performed directly on a bacterium. Finally, they isolated modified and unmodified YCpMmyc1.1 clones from the yeast cells and they unsuccessfully attempted transplanting them into M. capricolum. They then tried again after either artificially methylating the clones or by silencing the restriction endonuclease found in the new host cells in order to prevent DNA degradation. This last attempt they claimed to have provided positive results and showed successful transplantation of the M. mycoides into M. capricolum cells.
Figures

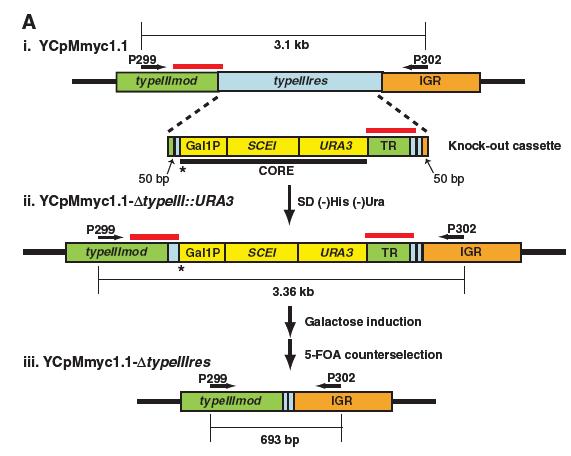
Figure 1A.
This figure is a schematic representation of the methodology followed to obtain YCpMmyc1.1 with a seamless deletion of the Type III restriction endonuclease (typeIIIres). They replaced the endonuclease with a knockout that was comprised of the URA3 marker, the Gal1 promoter (Gal1P), the SCEI endonuclease gene, and a series of tandem repeats (TR) (i). The arrow coupled with the “SD(-)His(-)Ura” represents their screening for cells that could grow on (-)His and (-)Ura medium which would mean they had incorporated the knock-out cassette in place of the endonuclease gene. In the (ii) part you can see a schematic of the YCpMmyc1.1 with the typeIIIres replaced by the cassette. The Galactose Induction arrow refers to the process by which they induced the production of the SCEI product through the Gal1 promoter, which would cleave the DNA at the asterisk and induce homologous recombination between the tandem repeats indicated by the red band. Finally, in order to select for the cells that underwent the seamless deletion of the knock-out (iii) they treated them with 5-fluoorotic acid, which targets the URA3 gene (5-FOA counterselection).
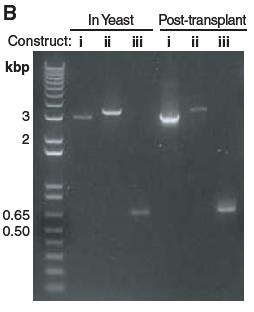
Figure 1B.
In this figure the authors are providing evidence to verify if the YCpMmyc1.1 of the transformed yeast actually underwent the seamless deletion. In order to achieve that, they performed PCR using primers P299 and P302 (represented by the black arrows seen in Figure 1A. on top of the DNA) to amplify the area of the restriction enzyme gene and its flanking DNA. They amplified DNA both from the yeast cells (In Yeast) and from transplanted M. capricolum cells (Post-Transplant). On the far left of the gel, you can see the molecular weight lane. In the (i) lanes they ran they amplified portion of the control unmodified YCpMmyc1.1 and it has the predicted size (3.1kb) as seen in Figure 1A. In the (ii) lanes they ran the YCpMmyc1.1 clones that have the knock-out cassette instead of the typeIIIres gene and once again the size of the amplicon is the predicted one (3.36kb). Finally, in the (iii) lane they ran the PRC product from the clones that had the cassette deleted and both bands are at the right size (693bp).
Table 1.
In this table the authors summarize the results of their attempts to transplant the wild-type M. capricolum and the strain with the silenced restriction endonuclease, M. capricolum RE (-). They used tetracycline to screen for the cells that took up the M. mycoides genome and the tetracycline-resistance gene found in the yeast vector. When the yeast strain was left untreated (no methylation) it was only taken up by the cells that had their restriction enzyme silenced (red square). Similarly, when the YCpMmyc1.1 transcripts were mock-methylated, treated the same as the methylated ones but there was no methyltransferase added, they were only successfully transplanted into the M. capricolum RE (-) and not the wild-type (red square). However, whenever the YCpMmyc1.1 was methylated with any of the three different methods (M. capricolum and M. mycoides extracts and the M. mycoides purified methylases) both the wild-type and the mutant strains were successfully transplanted. Finally, the checked to see if the modified versions of the YCpMmyc1.1 (with the knock-out cassette, with deleted typeIIIres gene and with 500kb deletion) could also be transplanted. The numbers show that with the exception of the 500kb deletion strain, which lacks essential genes, all the other versions of the genome were successfully transplanted.
.
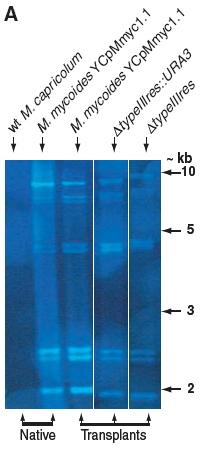
Figure 2A
This figure is a Southern blot with which they are providing evidence to support their claim of having successfully transplanted the M. capricolum by probing the DNA of each transplanted and untransplanted strain with M. mycoides-specific IS1296 element. In the first two lanes there are the negative and the positive controls respectively. In the first lane, there is the wild-type M. capricolum genomic DNA from unaffected cells and in the second lane the native M. mycoides genomic DNA from the YCpMmyc1.1 construct both digested with Hind III. The lack of bands in the first lane indicates that the probe only binds to the M. mycoides genomic DNA and shows distinct band patterns for M. mycoides DNA. The last three are the experimental lanes, containing digested DNA from transplanted cells with one of the three types M. mycoides genomic DNA. All of the experimental lanes match the banding pattern of the native M. mycoides’ lane suggesting that it was successfully transplanted into the M. capricolum cells.
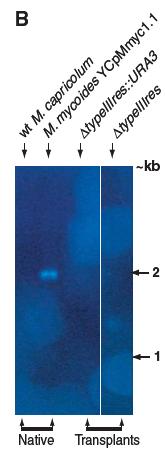
Figure 2B
In this figure the authors, using Southern blot analysis, provide evidence for the deletion of the typeIIIres gene in the YCpMmyc1.1 genomes found in the transplanted cells by Southern blot analysis using. The genomic DNA ran in the gel was digested with Eco RV and the probe used was the typeIIIres gene sequence. Similar to Figure 2A, the first two lanes are the negative and the positive controls, namely the wild type M. capricolum and M. mycoides with the YCpMmyc1.1 construct. In the second to last lane there is the DNA of the cells transplanted with the knock-out cassette version of the YCpMmyc1.1 and in the last lane is the DNA from the cells transplanted with the deleted typeIIIres version. As it was expected, only the native M. mycoides cells have the typeIIIres in their DNA.
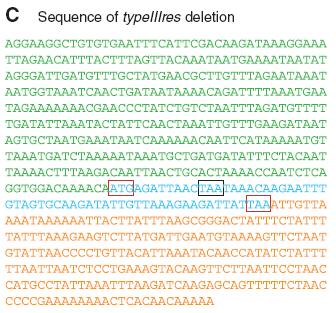
Figure 2C.
Lastly, the authors in order to verify the absence of the typeIIIgene in the transplanted cells they sequenced the specific region of one of the strains. The color coding of the sequence is the same as the one in the gene map seen in the Figure 1A, so the green portion is the typeIIImod portion, the blue is the small remaining part of the typeIIIres, and the orange part is the IGR. This sequencing shows that the core of the typeIIIres gene is missing from the genome of the transplanted cells. The remaining portion is flanked by start and stop codons as it is indicated by the red and black boxes which would render this DNA untranslatable.
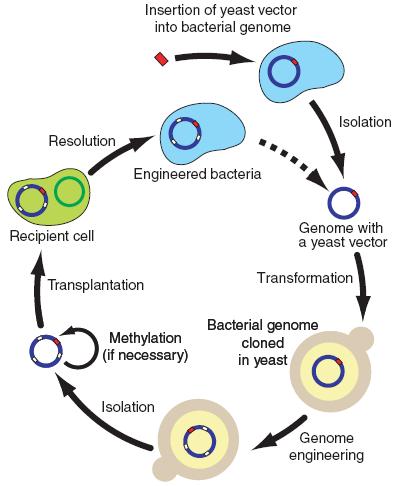
Figure 3.
This figure is a simple graphical representation of the steps followed to obtain a bacteria with cloned and engineered DNA starting with inserting a yeast vector in the bacterial genome, then transforming the bacterial genome in the yeast, performing any desired modifications to the DNA, methylating the bacterial genome and transplanting into the new bacterial cells.
Paper Critique
Overall the paper successfully presents evidence to support the effectiveness of this newly developed technique. However, there are parts that I believe could be improved upon. First of all, I believe that the part about the Mycoplasma genitalium in the first paragraph does not contribute much to their overall argument and it personally left me confused as to why they spend such little time on something they mentioned this early in their paper. Also, as a reader of scientific papers I prefer when the authors mention the greater implications of their results earlier in the paper instead of in the very end so as to create a better framework with which to read the rest of the paper. One of the things that confused me about their methodology and I would have liked them to explain more thoroughly was why they chose to transplant the modified genome from the yeast into a new related species and not the original species from which the genome was obtained.
If we look specifically at the figures of the paper, I believe that the gene maps in Figure 1A at first seemed to be condensed with a lot of information and acronyms but with closer readings it clearly explains the steps they followed to obtain the seamless deletion of the typeIIIres gene. Similarly, Figure 1B is very clear and the bands seen on the gel undoubtedly support their original hypothesis and fit the predicted size of the transcripts.
Table 1 manages to present in an organized manner their result and also they do not try to unreasonably interpret any of their results, as seen in this table. It is clear that there is a distinct difference between the absence of blue colonies seen wild-type M. capricolum, the 37 blue colonies of the mutated strain and the positive numbers seen after the methylation treatment. This difference is enough for them to make their case that silencing the restriction endonuclease in the recipient cells and the methylating treatment is sufficient for successfully transplantion of the YCpMmyc1.1.
In Figures 2A and 2B, the point that the authors are trying to make would have been a lot stronger if they probed the digested genomic DNA in all the lanes with a generic sequence that would served as a loading control. The way the blots are right now is a little ambiguous because the reader cannot be certain if the negative results seen in the lanes are because of actual differences or because of complete lack of DNA in that particular lane. Also, as a different control I would have like to see them probe the same digestic DNA with sequences specific to M. capricolum and to yeast in order to ensure that the DNA in the found in the transplanted cells is exclusively from the M. mycoides.
Finally, Figure 3 neatly concludes and summarized the main points of their newly developed method. However, I believe, as I mentioned above for figure 2A-C, fails to provide sufficient evidence in order to support the part that they refer to in this last figure as “Resolution”. In other words, I believe that with the data that they are providing us they have not completely shown that the DNA present in the transplanted cells is exclusively from the M. mycoides species.
Future Experiments
One of the points made in Table 1 that is not touched upon at all in their discussion and looks like an interesting point from which future experiment can jumpstart from is the obvious difference in the number of transplant colonies produced by the mutant strain of the M. capricolum. The number of colonies range from a total of 15 to 52 blue colonies indicating that there might be other factors that make some of the transplantation attempts more or less efficient. Since, this is such a newly developed technique I believe trying to figure the best and most efficient conditions under which it should be performed to produce a large number of colonies. Understanding the biological mechanisms underlying this method will also allow researchers to safely use it and in pathological research for humans
As it is mentioned in the paper, this new method has just opened the doors to manipulating the bacterial genome in ways previously unimaginable, using the very well defined yeast model and its already existing genetic techniques. Using this method can definitely allow medical researchers to explore more the biological mechanisms that drive a number of different pathogenic bacteria, including the mycoides. For instance, knocking-out specific genes in the bacterial genome, in exactly the same way as it was done in this paper, it could allow researchers to uncover the specific genes that make a bacterium pathogenic and specifically target those genes when looking for treatment methods.
References
Lartigue C, Vashee S, Algire MA, Chuang R, Benders GA, Ma L, Noskov VN, Denisova EA, Gibson DG, Assad-Garcia N, Alperovich N, Thomas DW, Merryman C, Hutchison CA, Smith HO, Venter JC, Glass JI. Creating Bacterial Strains from Genomes That Have Been Cloned and Engineered in Yeast. Science 2009; 325: 1693-1696. Abstract
Ilias Theodorou's Home Page Molecular Biology Home Page
For any questions or comments please email iltheodorou@davidson.edu